


Next: Artificial Neural Systems
Up: Clustering Algorithms
Previous: Robust -medoids
Contents
Nearest-neighbor clustering is an agglomerative single-link clustering
technique. Starting with each sample as a singleton cluster, at each
stage, the closest pair of clusters is merged. After 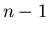 stages,
only one cluster remains and the resulting binary tree can be cut at
the desired (or user specified) depth to yield a specific
partitioning. The central design choice is the definition of cluster
distance:
stages,
only one cluster remains and the resulting binary tree can be cut at
the desired (or user specified) depth to yield a specific
partitioning. The central design choice is the definition of cluster
distance:
- Minimum distance
- (single-link).
The distance of two clusters equals the minimum distance of all pairs
of points taken one from each cluster. This definition yields an
algorithm similar to the Minimum Spanning Tree (MST) algorithm and is
susceptible to `bridging': a line of points connecting clusters may
cause their unintended merge.
- Maximum distance
- (complete-link).
Conversely, using the maximum pair-wise distance prefers spherical and
compact clusters and minimizes cluster diameter.
- Average distance.
- Balancing this trade-off one might
also choose the average pair-wise distance as the cluster
distance.
- Mean distance.
- An efficient method is to compute the mean of
each cluster and define cluster distance as the distance between the
two clusters means. Then, not all pairs' distances have to be
computed.
However, the 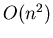 to
to 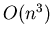 complexity of nearest-neighbor
clustering and the greedy nature without any backtracking are
significant drawbacks.
complexity of nearest-neighbor
clustering and the greedy nature without any backtracking are
significant drawbacks.



Next: Artificial Neural Systems
Up: Clustering Algorithms
Previous: Robust -medoids
Contents
Alexander Strehl
2002-05-03
![]() stages,
only one cluster remains and the resulting binary tree can be cut at
the desired (or user specified) depth to yield a specific
partitioning. The central design choice is the definition of cluster
distance:
stages,
only one cluster remains and the resulting binary tree can be cut at
the desired (or user specified) depth to yield a specific
partitioning. The central design choice is the definition of cluster
distance: